Anomalous Difference Map
In Basics, we loaded a room-temperature dataset that was collected at ~6550 eV of tetragonal HEWL. We then computed the \(CC_{1/2}\) and \(CC_{anom}\) for this data in Merging Statistics and observed that there was significant anomalous signal. Let’s now use that data to generate an anomalous difference map based on the anomalous scattering from the native sulfur atoms.
[1]:
import reciprocalspaceship as rs
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("notebook", font_scale=1.3)
import numpy as np
[2]:
print(rs.__version__)
0.10.3
[3]:
# Load data and extract relevant columns
refltable = rs.read_mtz("data/HEWL_SSAD_24IDC.mtz")
refltable = refltable[["I(+)", "SIGI(+)", "I(-)", "SIGI(-)", "N(+)", "N(-)"]]
refltable.head()
[3]:
| I(+) | SIGI(+) | I(-) | SIGI(-) | N(+) | N(-) | |||
|---|---|---|---|---|---|---|---|---|
| H | K | L | ||||||
| 0 | 0 | 4 | 661.29987 | 21.953098 | 661.29987 | 21.953098 | 16 | 16 |
| 8 | 3229.649 | 105.980934 | 3229.649 | 105.980934 | 16 | 16 | ||
| 12 | 1361.8672 | 43.06085 | 1361.8672 | 43.06085 | 16 | 16 | ||
| 16 | 4124.393 | 196.89108 | 4124.393 | 196.89108 | 8 | 8 | ||
| 1 | 0 | 1 | 559.33685 | 8.6263 | 559.33685 | 8.6263 | 64 | 64 |
[4]:
print(f"Number of reflections: {len(refltable)}")
Number of reflections: 12542
Background
Since this dataset was collected at a single wavelength, we can compute an anomalous difference map from the anomalous structure factor amplitudes, \(|F_{A}|\), and their phase shifts, \(\alpha\), relative to the computed phases from an isomorphous structure of tetragonal HEWL. We will compute \(|F_{A}|\) based on the following:
\begin{equation*} |F_{A}| \propto \Delta_{\mathrm{ano}} = |F_{HKL}| - |F_{\overline{HKL}}| \end{equation*}
We will then use a model refined from this data to obtain the phases, \(\phi_c\), which can be used to determine the phases for the anomalous contribution, \(\phi_A\), using the phase shift, \(\alpha\):
\begin{equation*} \phi_A = \phi_c - \alpha \end{equation*}
Since this is a SAD experiment, we can assume \(\alpha\) is 90˚ when \(\Delta_{\mathrm{ano}}\) is negative and 270˚ when it is positive. This formalism is based on Thorn and Sheldrick, J Appl. Cryst. (2011).
Computing Structure Factor Amplitudes
The dataset being used is the direct output from running scaling and merging in AIMLESS. As a first processing step, we need to convert the observed merged intensities, \(I(+)\) and \(I(-)\), into observed structure factor amplitudes, \(F(+)\) and \(F(-)\).
[5]:
# Stack intensities from 2-column anomalous to 1-column format
stacked = refltable.stack_anomalous()
stacked = stacked.loc[stacked["N"] > 0]
stacked.sort_index(inplace=True)
stacked.head()
[5]:
| I | SIGI | N | |||
|---|---|---|---|---|---|
| H | K | L | |||
| -45 | -10 | -1 | 6.185645 | 2.932488 | 4 |
| -9 | -2 | 27.028767 | 3.8457258 | 4 | |
| -1 | 3.0018542 | 2.6649861 | 4 | ||
| -8 | -3 | -0.9806365 | 2.7741797 | 4 | |
| -2 | 12.085027 | 3.0270035 | 4 |
In order to get structure factor amplitudes, we must first account for mean intensities that are negative due to background subtraction. We will use a method based on the Bayesian approach first proposed by French and Wilson to ensure that all intensities are strictly positive. This method is implemented in rs.algorithms.scale_merged_intensities().
[6]:
scaled = rs.algorithms.scale_merged_intensities(stacked, "I", "SIGI",
mean_intensity_method="anisotropic")
[7]:
scaled.head()
[7]:
| I | SIGI | N | dHKL | CENTRIC | FW-I | FW-SIGI | FW-F | FW-SIGF | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H | K | L | |||||||||
| -45 | -10 | -1 | 6.185645 | 2.932488 | 4 | 1.7194302 | False | 6.1471047 | 2.7739754 | 2.4032679 | 0.6094333 |
| -9 | -2 | 27.028767 | 3.8457258 | 4 | 1.7217722 | False | 26.716537 | 3.8457258 | 5.1551414 | 0.37557602 | |
| -1 | 3.0018542 | 2.6649861 | 4 | 1.7271528 | False | 3.5474997 | 2.1485696 | 1.7800175 | 0.6156602 | ||
| -8 | -3 | -0.9806365 | 2.7741797 | 4 | 1.7197413 | False | 1.8468517 | 1.4775624 | 1.2411455 | 0.5535428 | |
| -2 | 12.085027 | 3.0270035 | 4 | 1.7287054 | False | 11.893286 | 3.02595 | 3.4186602 | 0.45392564 |
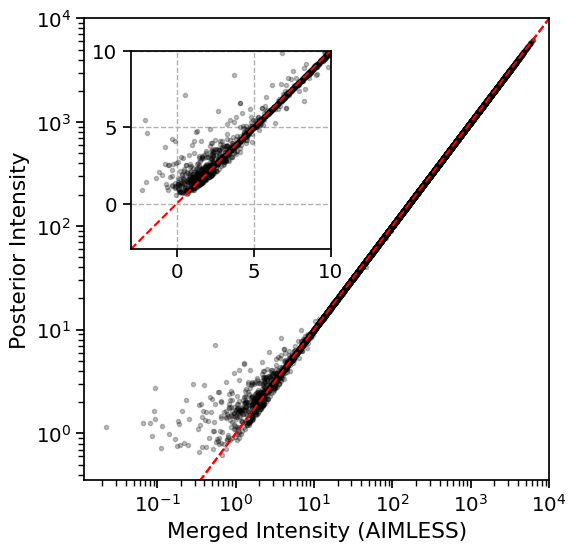
French-Wilson scaling leaves large intensities relatively unchanged, but rescales negative and small intensities to be positive by imposing Wilson’s distribution as a prior:
[8]:
x = np.linspace(-3, 10000, 2)
plt.figure(figsize=(6, 6))
ax = plt.gca()
plt.plot(x, x, '--r', scalex=False, scaley=True)
plt.plot(scaled["I"].to_numpy(), scaled["FW-I"].to_numpy(), "k.", alpha=0.25)
plt.plot(plt.xlim(), plt.xlim(), '--r', scalex=False, scaley=True)
plt.xlabel("Merged Intensity (AIMLESS)")
plt.ylabel("Posterior Intensity")
plt.xscale("log")
plt.yscale("log")
plt.xlim(right=1e4)
plt.ylim(top=1e4)
# Inset
axins = ax.inset_axes([0.1, 0.5, 0.43, 0.43])
axins.plot(scaled["I"].to_numpy(), scaled["FW-I"].to_numpy(), "k.", alpha=0.25)
axins.plot(x, x, '--r')
axins.set_xlim(-3, 10)
axins.set_ylim(-3, 10)
axins.grid(linestyle="--")
plt.show()
[9]:
# Remove extra columns
scaled = scaled[["FW-F", "FW-SIGF", "N"]]
# "Unstack" anomalous data from one-column to two-column format
anom = scaled.unstack_anomalous(["FW-F", "FW-SIGF", "N"]).dropna()
[10]:
anom.head()
[10]:
| FW-F(+) | FW-SIGF(+) | N(+) | FW-F(-) | FW-SIGF(-) | N(-) | |||
|---|---|---|---|---|---|---|---|---|
| H | K | L | ||||||
| 0 | 0 | 4 | 25.7036 | 0.4273095 | 16 | 25.7036 | 0.4273095 | 16 |
| 8 | 56.794975 | 0.93358195 | 16 | 56.794975 | 0.93358195 | 16 | ||
| 12 | 36.885887 | 0.5840336 | 16 | 36.885887 | 0.5840336 | 16 | ||
| 16 | 64.02966 | 1.5395098 | 8 | 64.02966 | 1.5395098 | 8 | ||
| 1 | 0 | 1 | 23.647089 | 0.18242109 | 64 | 23.647089 | 0.18242109 | 64 |
[11]:
# Compute differences
dF = np.abs(anom["FW-F(+)"] - anom["FW-F(-)"])
sigDF = np.sqrt(anom["FW-SIGF(+)"]**2 + anom["FW-SIGF(-)"]**2)
anom["ANOM"] = rs.DataSeries(dF, dtype="SFAmplitude")
anom["SigANOM"] = rs.DataSeries(sigDF, dtype="Stddev")
[12]:
anom.head()
[12]:
| FW-F(+) | FW-SIGF(+) | N(+) | FW-F(-) | FW-SIGF(-) | N(-) | ANOM | SigANOM | |||
|---|---|---|---|---|---|---|---|---|---|---|
| H | K | L | ||||||||
| 0 | 0 | 4 | 25.7036 | 0.4273095 | 16 | 25.7036 | 0.4273095 | 16 | 0.0 | 0.60430694 |
| 8 | 56.794975 | 0.93358195 | 16 | 56.794975 | 0.93358195 | 16 | 0.0 | 1.3202842 | ||
| 12 | 36.885887 | 0.5840336 | 16 | 36.885887 | 0.5840336 | 16 | 0.0 | 0.82594824 | ||
| 16 | 64.02966 | 1.5395098 | 8 | 64.02966 | 1.5395098 | 8 | 0.0 | 2.1771955 | ||
| 1 | 0 | 1 | 23.647089 | 0.18242109 | 64 | 23.647089 | 0.18242109 | 64 | 0.0 | 0.25798237 |
Phasing the Anomalous Difference Map
Below, we will compute the necessary phase shifts to go from the phases of a \(2 F_o - F_c\) map to the phases associated with the anomalous difference structure factors. Although this model was refined to this data in PHENIX, the phases from any isomorphous structure could have been used to obtain a reasonable map.
[13]:
ref = rs.read_mtz("data/HEWL_refined.mtz")
[14]:
# Find common HKL indices
hkls = anom.index.intersection(ref.index).sort_values()
anom = anom.loc[hkls]
As mentioned in Background, we can compute the anomalous phases as follows:
\begin{equation*} \phi_A = \phi_c - \alpha \end{equation*}
where \(\alpha\) is 90˚ when \(\Delta_{\mathrm{ano}}\) is negative and 270˚ when it is positive.
[15]:
alpha = 90 + 180*(anom["FW-F(+)"] >= anom["FW-F(-)"])
anom["PHANOM"] = ref.loc[hkls, "PH2FOFCWT"] + alpha
anom["PHANOM"] = rs.utils.canonicalize_phases(anom["PHANOM"]).astype("Phase")
Viewing the Map
Since we have structure factor amplitudes, anom["ANOM"], and phases, anom["PHANOM"], for the anomalous structure factors, we can now view the anomalous difference map. This can be done by writing out an MTZ file, and loading it into COOT, PyMOL, or any other molecular graphics package.
[16]:
anom.dtypes
[16]:
FW-F(+) FriedelSFAmplitude
FW-SIGF(+) StddevFriedelSF
N(+) MTZInt
FW-F(-) FriedelSFAmplitude
FW-SIGF(-) StddevFriedelSF
N(-) MTZInt
ANOM SFAmplitude
SigANOM Stddev
PHANOM Phase
dtype: object
[17]:
anom.write_mtz("data/anomdiff.mtz")
Looking at the anomalous difference map on the refined structure, we can see positive difference density on all of the sulfurs in HEWL (shown below in purple):
[18]:
%%html
<center>
<iframe src="https://rs-station.github.io/reciprocalspaceship/data/anomdiff/anomdiff.html#zoom=25", width=600, height=600></iframe>
<br>Anomalous difference map from HEWL crystal overlayed with refined model (PDB: 7L84). Map is contoured at +5σ.
</center>
Anomalous difference map from HEWL crystal overlayed with refined model (PDB: 7L84). Map is contoured at +5σ.