Basics
reciprocalspaceship provides methods for reading and writing MTZ files, and can be easily used to join reflection data by Miller indices. We will demonstrate these uses by loading diffraction data of tetragonal hen egg-white lysozyme (HEWL).
[1]:
import reciprocalspaceship as rs
print(rs.__version__)
0.9.9
This diffraction data was collected at the Sector 24-ID-C beamline at NE-CAT at APS. Diffraction images were collected at ambient room temperature (295K), and low energy (6550 eV) in order to collect native sulfur anomalous diffraction for experimental phasing. The diffraction images were processed in DIALS for indexing, geometry refinement, and spot integration, and scaling and merging was done in
AIMLESS. This data reduction yielded an MTZ file that is included in the data/ subdirectory. Here, we will load the MTZ file and inspect its contents.
Loading reflection data
Reflection tables can be loaded using the top-level function, rs.read_mtz(). This returns a DataSet object, that is analogous to a pandas.DataFrame.
[2]:
refltable = rs.read_mtz("data/HEWL_SSAD_24IDC.mtz")
type(refltable).__name__
[2]:
'DataSet'
This reflection table was produced directly from AIMLESS, and contains several different data columns:
[3]:
refltable.head()
[3]:
| FreeR_flag | IMEAN | SIGIMEAN | I(+) | SIGI(+) | I(-) | SIGI(-) | N(+) | N(-) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H | K | L | |||||||||
| 0 | 0 | 4 | 14 | 661.29987 | 21.953098 | 661.29987 | 21.953098 | 661.29987 | 21.953098 | 16 | 16 |
| 8 | 4 | 3229.649 | 105.980934 | 3229.649 | 105.980934 | 3229.649 | 105.980934 | 16 | 16 | ||
| 12 | 6 | 1361.8672 | 43.06085 | 1361.8672 | 43.06085 | 1361.8672 | 43.06085 | 16 | 16 | ||
| 16 | 19 | 4124.393 | 196.89108 | 4124.393 | 196.89108 | 4124.393 | 196.89108 | 8 | 8 | ||
| 1 | 0 | 1 | 16 | 559.33685 | 8.6263 | 559.33685 | 8.6263 | 559.33685 | 8.6263 | 64 | 64 |
[4]:
print(f"Number of reflections: {len(refltable)}")
Number of reflections: 12542
Internally, each of these data columns is stored using a custom dtype that was added to the conventional pandas and numpy datatypes. This enables DataSet reflection tables to be written back to MTZ files. There is a dtype for each of the possible datatypes listed in the MTZ file specification.
[5]:
refltable.dtypes
[5]:
FreeR_flag MTZInt
IMEAN Intensity
SIGIMEAN Stddev
I(+) FriedelIntensity
SIGI(+) StddevFriedelI
I(-) FriedelIntensity
SIGI(-) StddevFriedelI
N(+) MTZInt
N(-) MTZInt
dtype: object
Additional crystallographic metadata is read from the MTZ file and can be stored as attributes of the DataSet. These include the crystallographic spacegroup and unit cell parameters, which are stored as gemmi.SpaceGroup and gemmi.UnitCell objects.
[6]:
refltable.spacegroup
[6]:
<gemmi.SpaceGroup("P 43 21 2")>
[7]:
refltable.cell
[7]:
<gemmi.UnitCell(79.3439, 79.3439, 37.8099, 90, 90, 90)>
Plotting reflection data
For illustrative purposes, let’s plot the \(I(+)\) data against the \(I(-)\) data
[8]:
%matplotlib inline
import matplotlib.pyplot as plt
[9]:
plt.figure(figsize=(6, 6))
plt.plot(refltable['I(+)'].to_numpy(), refltable['I(-)'].to_numpy(), 'k.', alpha=0.1)
plt.xlabel("I(+)")
plt.ylabel("I(-)")
plt.show()
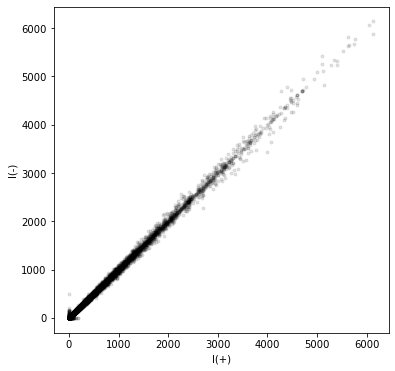
In the next example, we will investigate this anomalous signal in more detail.
Writing Reflection Data
It is also possible to write out MTZ files using DataSet.write_mtz(). This functionality depends on the correct setting of each column’s dtype.
[10]:
refltable.write_mtz("data/HEWL_SSAD_24IDC.mtz")