19 Mar 2025
Generative ML Approach to Electron Density Map Sharpening
Variational Autoencoding for Map Sharpening
Jacob Thornton
Map Sharpening
X-ray crystallography and Cryo-electron microscopy are important techniques to determine atomic-level information of biological molecules. They are widely used in many fields including structural biology, biochemistry, and biophysics. Therefore, the task of “map sharpening” – taking in either low-resolution electron density maps in crystallography or electrostatic potential maps in cryo-EM to create high-resolution ones – is a valuable area of research. Known as super-resolution in computer science, this class of algorithms increases the information content of low-resolution data. Our project focuses on this task of map sharpening, taking a 3-dimensional, low-resolution electron density map and outputting one with more detail. More detailed maps make it easier for structural biologists to interpret their data by modeling atoms into the density. Map sharpening has the potential to save researchers time and improve automated structure modeling. This task is often difficult, but has been made more feasible with machine learning algorithms.
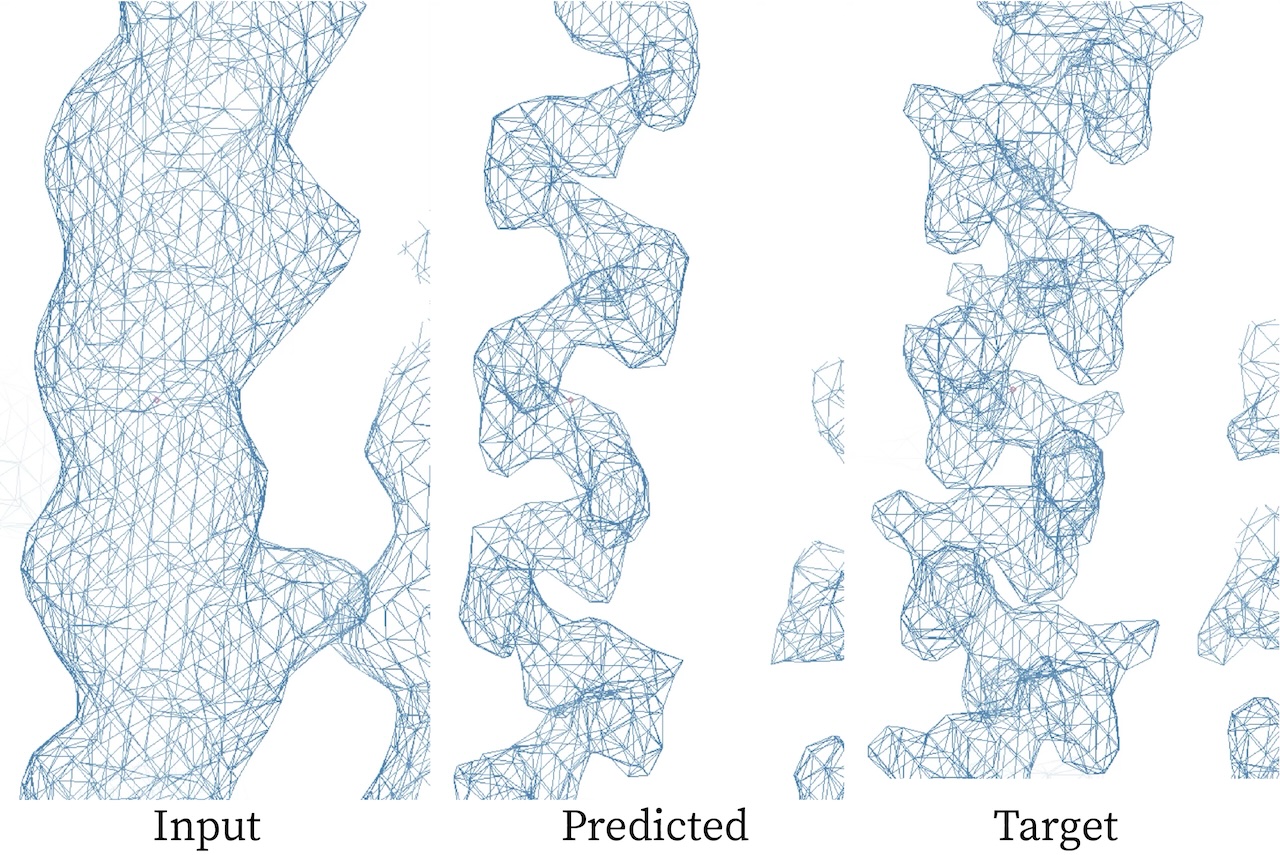 Example of map sharpening through machine learning
Example of map sharpening through machine learning
Generative Modeling
A number of successful machine learning models for map sharpening have been produced in recent years but there has yet to be a significant success of generative models for this task. With our research we aim to determine the feasibility of a variational autoencoder (VAE) as a model for electron density map sharpening. A VAE model is composed of an encoder and a decoder. The encoder encodes important information or features of the input map to a lower dimensional space called the latent space. This encoded vector of features is then scaled back up by the decoder to the size of the input to retrieve the sharpened map.
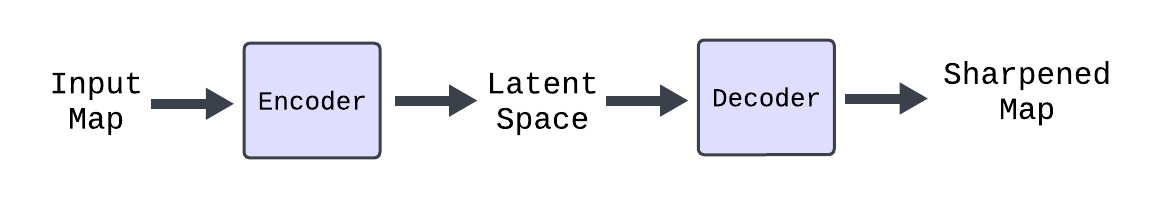 VAE architecture visualization
VAE architecture visualization
Results
A lot of progress was made toward building a VAE model actually capable of sharpening electron density maps. Although current results are short of performing super resolution, they have validated the use of a VAE in that our model is capable of encoding relevant information from a map to a latent space and then decoding that to something that resembles a density. That being said there is still a significant gap between our results and the threshold for super resolution as measured by the real space correlation coefficient of the produced maps on target maps in the validation set.
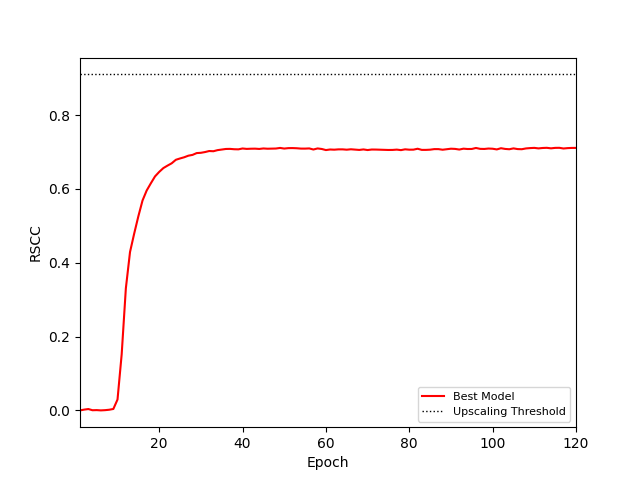 Validation real space correlation coefficient
Validation real space correlation coefficient
Next Steps
Next steps will involve further modifying the VAE architecture to improve model performance. This will consist primarily of increasing the power of the encoder to reduce input dimensionality before it is mapped into the latent space and secondarily of continuing to modify hyperparameters such as the learning rate of the model.