22 Aug 2024
Merging Small Molecule MicroED Data
Preprint Demonstrates using Careless in Small Molecule MicroED
Doris Mai
Small Molecule MicroED
Microcrystal electron diffraction (MicroED), also known as continuous rotational electron diffraction (cRED), or 3-dimensional electron diffraction (3DED), is an emerging technique for elucidating small molecule structures from micro- and nano-crystals. However, data processing in microED can be challenging and often requires merging datasets from multiple crystals due to limited rotation ranges in many transmission electron microscopes (TEMs) and higher noise in intensity measurements than conventional X-ray diffraction (XRD) experiments. A common practice in the field is to manually curate datasets and apply scaling programs such as XDS/XSCALE from rotational XRD, but this can be time-consuming and risks introducing human bias. In this preprint, we demonstrate that Careless can be used for merging small molecule microED data and investigate the impact of dataset curation.
Dataset Curation in Merging
We benchmark Careless with XDS/XSCALE on more than 10 cases of multi-crystal merging and compare the performance between using all datasets and manually curated datasets (curated=True). We also explore an extension to Careless (MC-Careless) to automate the curation of datasets. Specifically, an optimal weighting among datasets (\(w\)) is learned jointly with the scaling factors (\(K\)) and structure factor amplitudes (\(F\)). This weight modulates the effective uncertainty of the intensities (\(\sigma_I\)) from different crystals to account for the variability of data quality across datasets.
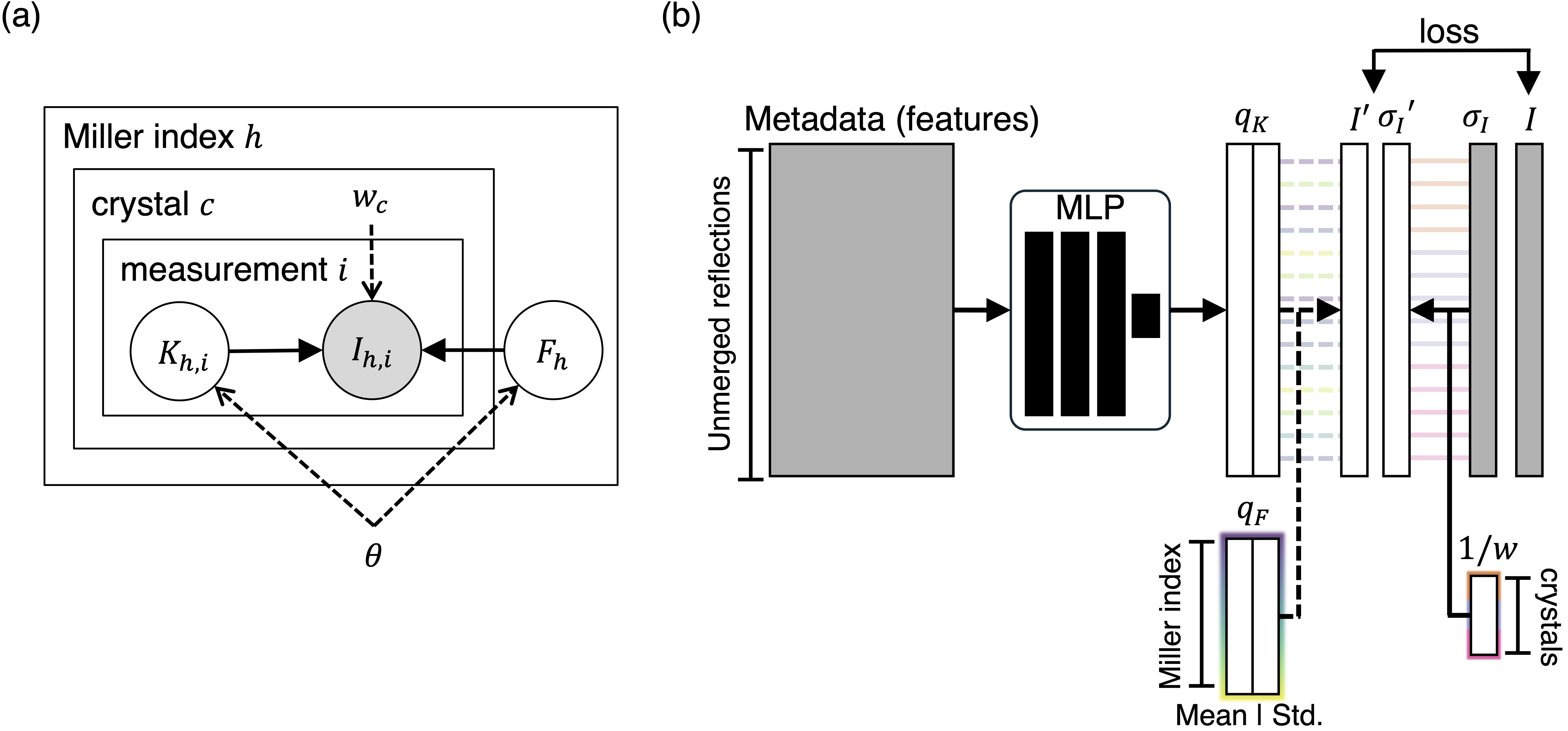 Schematic from the preprint of the multi-crystal weighting extension to Careless. (CC-BY-NC-ND license)
Schematic from the preprint of the multi-crystal weighting extension to Careless. (CC-BY-NC-ND license)
In the presented cases, Careless benefits from using all available data, and our multi-crystal weighting extension did not further improve the performance. Manual curation of datasets affects the \(CC_{1/2}\) in XDS/XSCALE and Careless but has marginal impact on the accuracy with respect to structure factor amplitudes calculated from reference structures and on ab initio phasing outcomes.
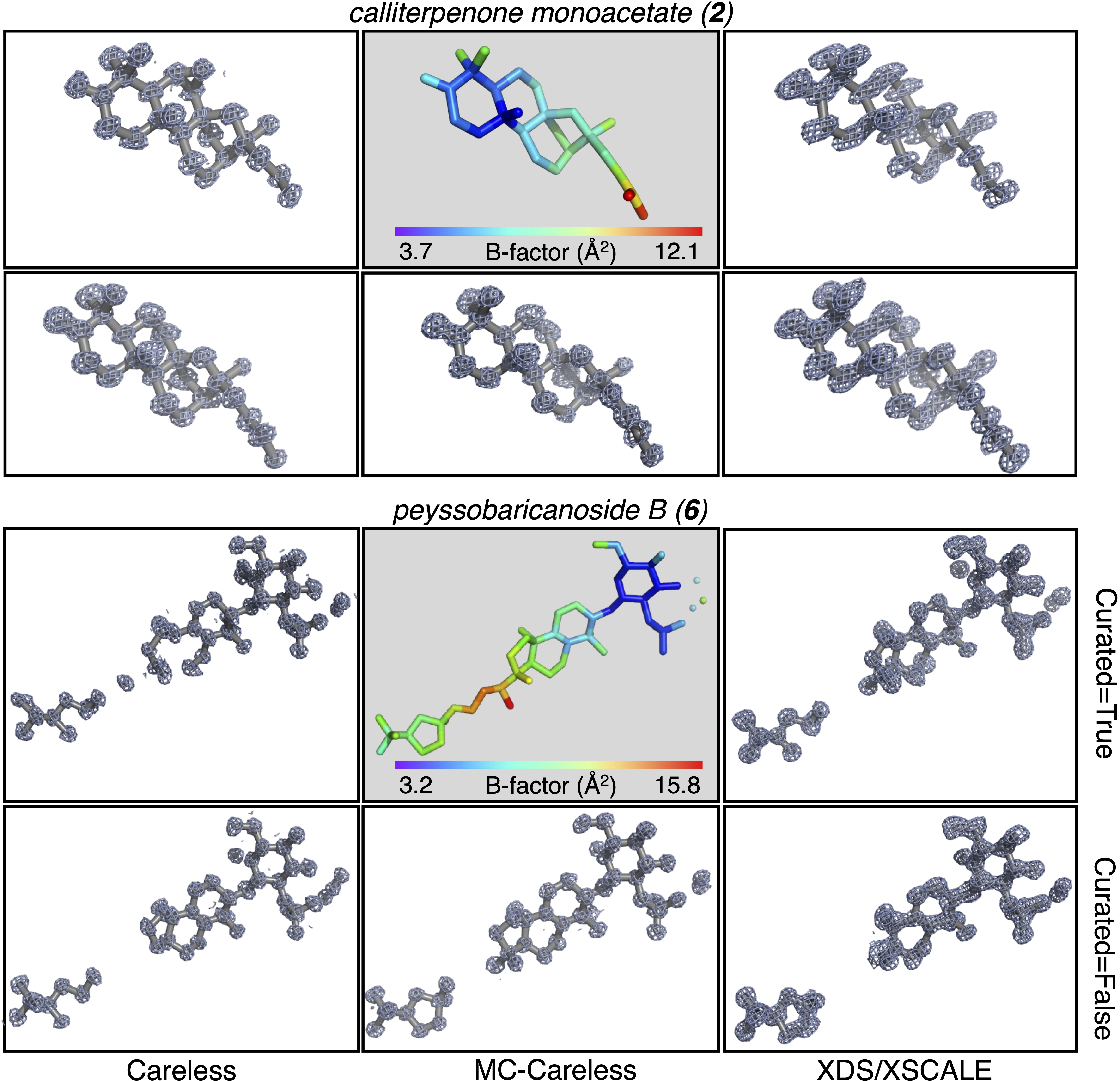 Examples from the preprint of ab initio phasing outcomes from different merging protocols. (CC-BY-NC-ND license)
Examples from the preprint of ab initio phasing outcomes from different merging protocols. (CC-BY-NC-ND license)
Key points and Future Directions
- Careless can merge microED and small molecule crystallography data.
- The variational inference framework implemented in Careless is flexible for methods development as demonstrated in our MC-Careless.
- Additional exploration of the hyperparamter space and the Wilson prior might be helpful for challeging cases with pathologies.
- The common practice of manual dataset curation should be cautioned and deserves future investigation.