Installation
The easiest way to install laue-dials and its dependencies is using
Anaconda.
First we update and install the libmamba solver with
conda update -n base conda
conda install -n base conda-libmamba-solver
conda config --set solver libmamba
With Anaconda, we can then create and activate a custom environment for your install by running
conda create --name laue-dials
conda activate laue-dials
Now we are ready to install the main dependency and framework:
DIALS. After installing that, we can
install laue-dials using pip, as below:
conda install -c conda-forge dials
pip install laue-dials
All other dependencies will then be automatically installed for us, and we’ll be ready to analyze your first Laue data set! Reopen this notebook with the appropriate environment activated when ready.
Documentation for laue-dials can be found at
here, and
entering a command with no arguments on the command line will also print
a help page!
Introduction
In this notebook, we will process a time-resolved EF-X dataset.
The data is comprised of four passes of four timepoints each. Each of
the four electric-field timepoints is taken for a given phi angle,
then the crystal is rotated around the phi goniometer axis and then the
four timepoints are taken again. This is done over four passes,
c,d,e,f. The start angle and the step can be found in the below
table.
pass |
start phi angle (deg) |
rotation phi step (deg) |
|---|---|---|
c |
0 |
2 |
d |
92 |
2 |
e |
181 |
2 |
f |
361.5 |
1 |
At the end of processing, we would like four .mtz files, one for
each timepoint. We must run laue-dials on each timepoint
individually, then combine all passes for a given timepoint at the end.
We start with the off timepoint pass c only. Then, we will
analyze all sixteen passes in a single script, and combine the output
mtzs into a single mtz file.
Data processing will rely on images found in ./data that can be downloaded
on SBGrid and scripts found in
./scripts downloadable here.
Importing Data
We can use dials.import as a way to import the data files written at
experimental facilities into a format that is friendly to both DIALS
and laue-dials. Feel free to use any data set you’d like below, but
a sample time-resolved EF-X data
set has been uploaded to Zenodo
for your convenience, and this notebook has been tested using that
dataset.
First, we create two dictionaries, START_ANGLES and OSCS, that
respectively map the pass names to the start angles and rotation steps
(treated as oscillations in dials.import).
declare -A START_ANGLES=( ["c"]=0 ["d"]=92 ["e"]=181 ["f"]=361.5)
declare -A OSCS=( ["c"]=2 ["d"]=2 ["e"]=2 ["f"]=1)
Then, we import the files for the c,off images using
dials.import.
#this is the delay time.
TIME="off"
# this is the pass.
pass="c"
FILE_INPUT_TEMPLATE="data/e35${pass}_${TIME}_###.mccd"
# Import data into DIALS files
dials.import geometry.scan.oscillation=${START_ANGLES[$pass]},${OSCS[$pass]}\
geometry.goniometer.invert_rotation_axis=True \
geometry.goniometer.axes=0,1,0 \
geometry.beam.wavelength=1.04 \
geometry.detector.panel.pixel_size=0.08854,0.08854 \
input.template=$FILE_INPUT_TEMPLATE \
output.experiments=imported.expt
Getting an Initial Estimate
After importing our data, the first thing we need to do is get an initial estimate for the experimental geometry. Here, we’ll use some monochromatic algorithms from DIALS to help! This step can be tricky – failure can be due to several causes. In the event of failure, here are a few common causes:
The spotfinding gain is either too high or too low. Try looking at the results of
dials.image_viewer imported.expt strong.refland seeing if you have too many (or too few) reflections. Lower gain gives you more spots, but also more likely to give false positives.Supplying the space group or unit cell during indexing can be helpful. When supplying the unit cell, allow for some variation in the lengths of the axes, since the monochromatic algorithms may result in a slightly scaled unit cell depending on the chosen wavelength.
You may have intensities that need to be masked. These can come from bad panels or extraneous scatter. You can use
dials.image_viewer(described below) to create a mask file for your data, and then provide thespotfinder.lookup.mask="pixels.mask"command below to use that mask during spotfinding.
laue.find_spots imported.expt \
spotfinder.mp.nproc=8 \
spotfinder.threshold.dispersion.gain=0.3 \
spotfinder.filter.max_separation=10
CELL='"65.3,39.45,39.01,90.000,117.45,90.000"' #this is a unit cell of PDZ2 from PDB 5E11
laue.index imported.expt strong.refl \
indexer.indexing.known_symmetry.space_group=5 \
indexer.indexing.refinement_protocol.mode=refine_shells \
indexer.indexing.known_symmetry.unit_cell=$CELL \
indexer.refinement.parameterisation.auto_reduction.action=fix \
laue_output.index_only=False
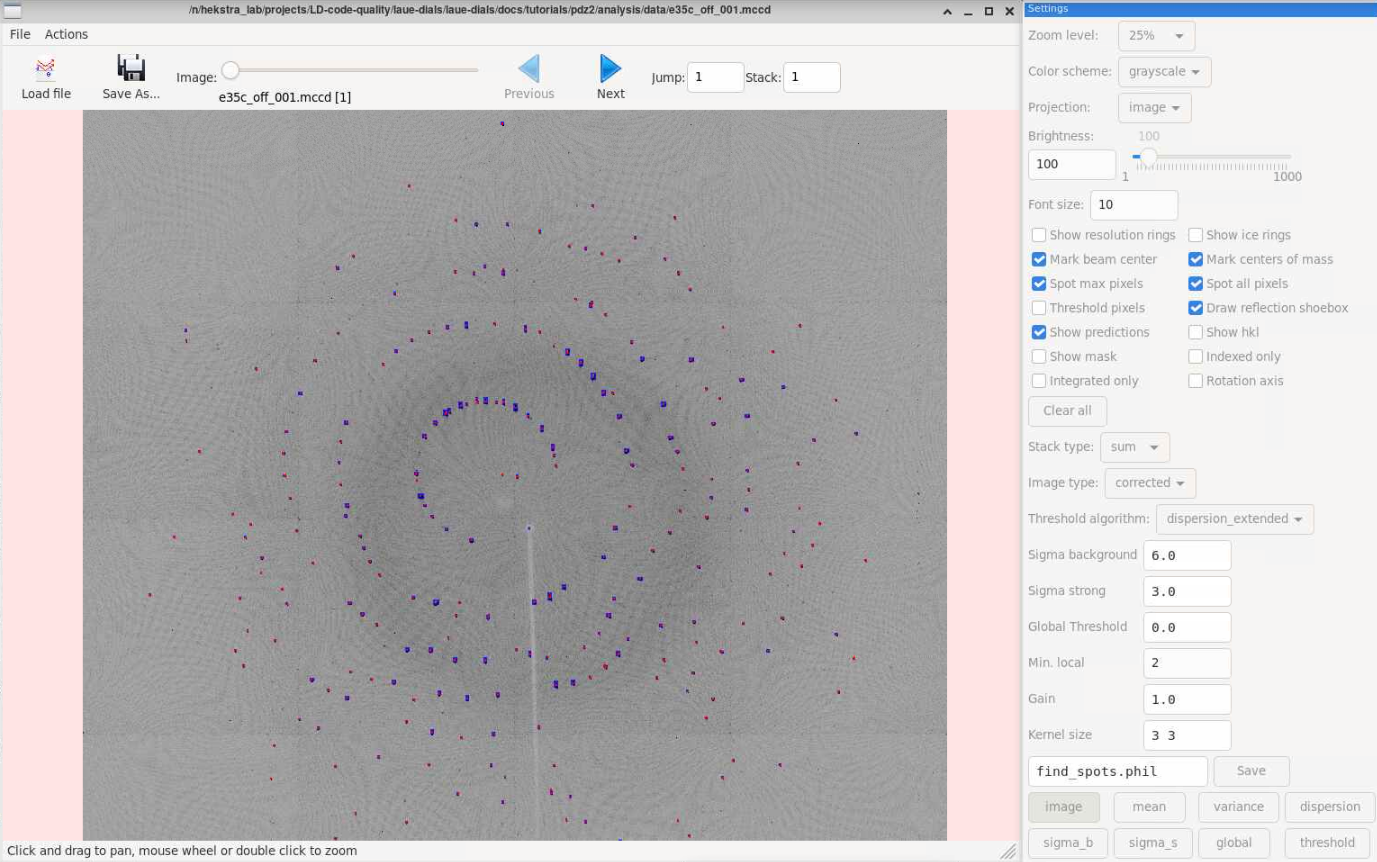
Viewing Images
Sometimes it’s helpful to be able to see the analysis data overlayed on
the raw data. DIALS has a utility for viewing spot information on the
raw images called dials.image_viewer. For example, the spotfinding
gain parameter can be tuned to capture more spots, but lowering it too
much finds nonexistent spots. To check this, we can use the image viewer
to see what spots were found on images. We need to provide an expt
file and a refl file – the imported.expt and strong.refl
files will do for checking spotfinding. This program also has utilities
for generating masks if they are needed. The red dots from the checkbox
“Mark centers of mass” are the spots found by laue.find_spots (which
in turn makes a call to dials.find_spots). These are best used for
judging whether you need to adjust the gain higher (for fewer spots) or
lower (for more) during spotfinding. You can find more details on the
image viewer in the DIALS tutorial
here.
dials.image_viewer imported.expt strong.refl
Making Stills
Here we will now split our monochromatic estimate into a series of
stills to prepare it for the polychromatic pipeline. There is a useful
utility called laue.sequence_to_stills for this.
NOTE: Do not use dials.sequence_to_stills, as there are data columns
which do not match between the two programs.
laue.sequence_to_stills monochromatic.*
Polychromatic Analysis
Here we will use four other programs in laue-dials to create a
polychromatic experimental geometry using our initial monochromatic
estimate. Each of the programs does the following:
laue.optimize_indexing assigns wavelengths to reflections and
refines the crystal orientation jointly.
laue.refine is a polychromatic wrapper for dials.refine and
allows for refining the experimental geometry overall to one suitable
for spot prediction and integration.
laue.predict takes the refined experimental geometry and predicts
the centroids of all strong and weak reflections on the detector.
laue.integrate then builds spot profiles and integrates intensities
on the detector.
laue.optimize_indexing stills.* \
output.experiments="optimized.expt" \
output.reflections="optimized.refl" \
output.log="laue.optimize_indexing.log" \
wavelengths.lam_min=0.95 \
wavelengths.lam_max=1.2 \
reciprocal_grid.d_min=1.7 \
nproc=8
laue.refine optimized.* \
output.experiments="poly_refined.expt" \
output.reflections="poly_refined.refl" \
output.log="laue.poly_refined.log" \
nproc=8 >> sink.log
To check the refinement quality, we check the spotfinding root-mean-square deviations (rmsds) as a function of image.
laue.compute_rmsds poly_refined.* refined_only=True show=True save=True
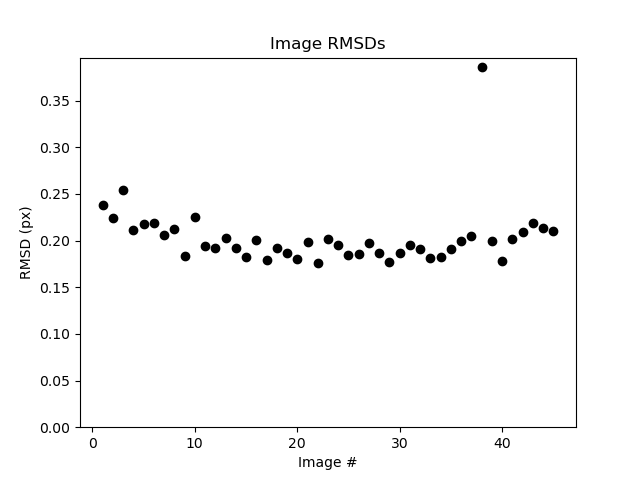
These rmsds look good.
Checking the Wavelength Spectrum
laue.plot_wavelengths allows us to plot the wavelengths assigned in
stored in a reflection table. The histogram of these reflections should
resemble the beam spectrum, so this is a good check to do at this time!
laue.plot_wavelengths poly_refined.refl refined_only=True save=True show=True
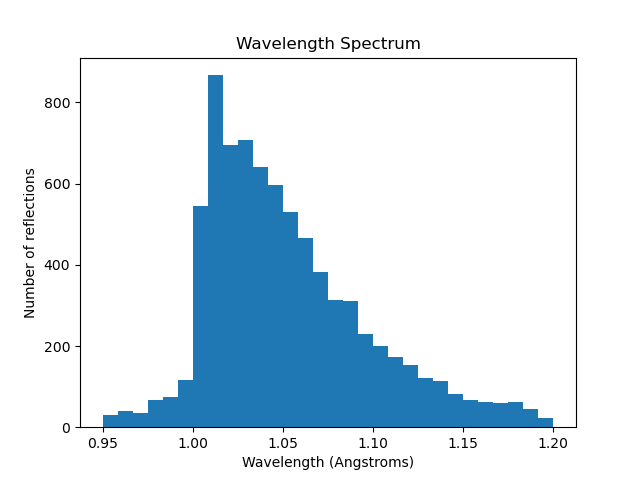
This is the expected wavelength profile, indicating successful wavelength assignment.
DIALS Reports
DIALS has a utility that gives useful information on various diagnostics
you may be interested in while analyzing your data. The program
dials.report generates an HTML file you can open to see information
and plots regarding the status of your analyzed data. You can run it on
any files generated by DIALS or laue-dials.
dials.report poly_refined.expt poly_refined.refl
Integrating Spots
Now that we have a refined experiment model, we can use laue.predict
and laue.integrate to get integrated intensities from the data. We
will predict the locations of all feasible spots on the detector given
our refined experiment model, and at each of those locations we will
integrate the intensities to get an mtz file that we can feed into
careless.
laue.predict poly_refined.* \
output.reflections="predicted.refl" \
output.log="laue.predict.log" \
wavelengths.lam_min=0.95 \
wavelengths.lam_max=1.2 \
reciprocal_grid.d_min=1.7 \
nproc=8
laue.integrate poly_refined.expt predicted.refl \
output.filename="integrated.mtz" \
output.log="laue.integrate.log" \
nproc=8
Processing and Combining All Passes
We have successfully integrated one of the sixteen image series. Let’s
now process the rest. For the off timepoints, we process as above.
Pass e has a different indexing solution (up to the C2 symmetry
operation -x,y,-z) and so we reindex pass e using
dials.reindex.
Our strategy for the 50ns,100ns,200ns timepoints is to
transfer the stills.expt geometry and then refine spot positions
that may have changed due to the electric field.
Using the attached ../scripts/one-pass-from_off.sh script which
contains all of the above bash code, we iterate over all of the
passes in the below cell. The below cell takes a while to run – we don’t
recommend to run this in the jupyter notebook. Instead, we recommend to
run it as a standalone parallel script, attached as
../scripts/process.sh. Either procedure will create a folder named
gain_0,3 containing subfolders of dials files for each pass. For
example, ../gain_0,3-from_stills/dials_files_d_100ns contains
dials files for pass d, timepoint 100ns.
declare -A START_ANGLES=( ["c"]=0 ["d"]=92 ["e"]=181 ["f"]=361.5)
declare -A OSCS=( ["c"]=2 ["d"]=2 ["e"]=2 ["f"]=1)
declare -A DELAY="off"
gain=0.3
for pass in c d e f
do
if [ pass == e ];then
sh scripts/one_pass-from_off.sh $pass $DELAY ${START_ANGLES[$pass]} ${OSCS[$pass]} $gain -x,y,-z >> sink.log
else
sh scripts/one_pass-from_off.sh $pass $DELAY ${START_ANGLES[$pass]} ${OSCS[$pass]} $gain x,y,z >> sink.log
fi
done
Once the off timepoint series finish, we process the remaining
timepoints.
declare -A START_ANGLES=( ["c"]=0 ["d"]=92 ["e"]=181 ["f"]=361.5)
declare -A OSCS=( ["c"]=2 ["d"]=2 ["e"]=2 ["f"]=1)
gain=0.3
for delay in "50ns" "100ns" "200ns"
do
for pass in "c" "d" "e" "f"
do
sh scripts/one_pass-from_off.sh $pass $delay ${START_ANGLES[$pass]} ${OSCS[$pass]} $gain x,y,z >> sink.log
done
done
Finally, we combine all .mtz files for passes of a single timepoint
using the attached scripts/expt_concat.py script. .mtz files can
be found in gain_0,3-from_stills/ld_0,3_mtzs.
python scripts/expt_concat.py 0.3
import reciprocalspaceship as rs
rs.read_mtz("gain_0,3/ld_0,3_mtzs/cdef_e35_off.mtz")
We expect a mtz file with about 350,000 reflections.
Conclusion
At this point, you now have integrated mtz files that you can pass
to careless for scaling and
merging. We provide an example careless script, found at
../scripts/careless-cdef-ohp-mlpw.sh. However, after all Laue-DIALS
files are printed out, ../scripts/reduce.sh can also be run for a
complete analysis.
Note that throughout this pipeline, you can use DIALS utilities like
dials.image_viewer or dials.report to check progress and ensure
your data is being analyzed properly. We recommend regularly checking
the analysis by looking at the data on images, which can be done by
dials.image_viewer FILE.expt FILE.refl.
These files are generally written as pairs with the same base name, with
the exception of combining imported.expt + strong.refl, or
poly_refined.expt + predicted.refl.
Also note that you can take any program and enter it on the command-line for further help. For example, writing
laue.optimize_indexing
will print a help page for the program. You can see all configurable parameters by using
laue.optimize_indexing -c.
This applies to all laue-dials command-line programs.
Congratulations! This tutorial is now over. For further questions, feel free to consult documentation or email the authors.
